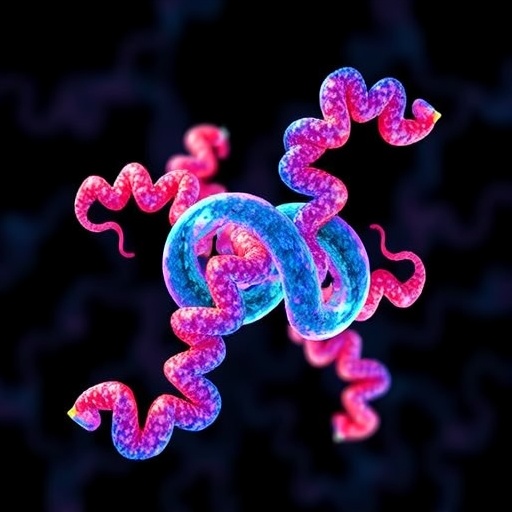
In a significant advancement for drug discovery, researchers have unveiled the potential of deep learning in protein-ligand docking, a crucial phase in understanding how small molecules interact with proteins. This breakthrough study, published in Nature Machine Intelligence, demonstrates how artificial intelligence (AI) can transform traditional computational methods, streamlining the drug development process and enhancing the efficacy of therapies.
Protein-ligand docking involves predicting how a ligand, a small molecule, or drug will orient itself when bound to a protein. Accurate predictions are essential, as they inform subsequent drug design stages, potentially saving time and resources. Historically, this process has required significant computational power and expertise in biochemistry. The advent of deep learning introduces a paradigm shift, enabling researchers to leverage AI to make precise predictions without the extensive traditional modeling.
The study, led by Morehead, Giri, and Liu, illustrates that deep learning offers unique advantages over conventional algorithms. By harnessing large datasets of known protein-ligand interactions, these models can uncover complex patterns that might escape human researchers. This capability promises to enhance the accuracy of docking predictions, which could substantially improve the efficiency of drug development pipelines.
Central to the success of these AI models is the quality and diversity of the datasets used for training. The researchers compiled extensive interaction data from publicly available databases, emphasizing that models lacking diverse training may yield unreliable results. The architecture employed, which includes convolutional neural networks (CNNs) and recurrent neural networks (RNNs), is adept at processing the three-dimensional structures of proteins and ligands. By learning from the intricacies of structural data, these networks generalize well to new interactions, enabling reliable predictions based solely on input structures.
A notable aspect of this research is its emphasis on interpretability—a common challenge in deep learning applications. The authors stress the importance of developing models that not only provide predictions but also elucidate the rationale behind them. This interpretability is crucial for fostering trust in AI-driven processes, particularly in drug discovery, where understanding binding predictions can guide further experimental verification.
The findings of this study hold promise for the future of precision medicine. By utilizing deep learning for protein-ligand docking, researchers can customize drug design according to individual patient profiles. This tailored approach not only enhances treatment efficacy but also seeks to reduce adverse side effects—an ongoing hurdle in pharmacology. The development of personalized therapies could revolutionize treatment strategies for complex diseases, such as cancer and autoimmune disorders.
Moreover, the implications of this research extend beyond small-molecule drug discovery. Insights gained from protein-ligand interactions may also inform the design of biologics, including antibodies and peptide-based therapies. Integrating deep learning models into the early design stages of these biological agents can streamline the identification of candidates likely to exhibit desired therapeutic effects.
A key takeaway from the study is the collaborative potential of AI in molecular biology. The authors clarify that while deep learning significantly enhances predictive accuracy, it should complement—not replace—human intuition and expertise. They advocate for hybrid approaches that merge AI’s strengths with researchers’ knowledge, promoting more robust drug discovery processes that ultimately benefit patient care.
While the excitement around these advancements is considerable, the researchers caution that it remains vital to validate AI predictions against experimental data. They highlight the importance of benchmark testing, where the accuracy of AI predictions is assessed against empirical results. This validation step is essential for establishing credibility within the scientific community, where rigorous data verification is foundational.
The timeline for realizing tangible benefits from these innovations may be shorter than previously expected. The integration of deep learning into protein-ligand docking offers pharmaceutical companies a pathway to accelerate their drug discovery timelines while exploring previously uncharted molecular structures. As more institutions adopt these technologies, the prospects for identifying novel therapeutics are set to increase dramatically.
In conclusion, the research by Morehead, Giri, and Liu showcases the transformative potential of deep learning in protein-ligand docking. By enhancing predictive accuracy and refining the drug discovery process, this study lays the groundwork for personalized medicine and innovative therapeutic strategies. As the scientific community increasingly embraces these cutting-edge technologies, remarkable advancements in tackling pressing health challenges can be anticipated.
As AI-driven research continues to evolve, the collaboration between computational and experimental scientists will be crucial. Navigating the complexities of molecular interactions through AI provides a promising roadmap for the future of medicine, where tailored treatments and efficient drug discovery processes become the standard. The intersection of deep learning and protein-ligand docking has the potential to redefine therapeutic development, empowering researchers to decode molecular interactions with unprecedented precision.
See also Korea’s Ministry of ICT Reveals Elite AI Teams Competing for National Foundation Model by 2027
Korea’s Ministry of ICT Reveals Elite AI Teams Competing for National Foundation Model by 2027 AI Models Exhibit Self-Preservation, Resisting Shutdown in 79% of Tests, Warns Palisade
AI Models Exhibit Self-Preservation, Resisting Shutdown in 79% of Tests, Warns Palisade Deep Learning Model Achieves 95% Accuracy in Automated Brain Tumor Detection
Deep Learning Model Achieves 95% Accuracy in Automated Brain Tumor Detection Researchers Giovanini and Moura Unveil Deep Learning Method for Indoor Sports Player Tracking
Researchers Giovanini and Moura Unveil Deep Learning Method for Indoor Sports Player Tracking