Researchers at Uppsala University have made significant strides in molecular modeling by applying machine learning techniques to more accurately predict molecular electrostatic potentials (MEP). The team, consisting of Kadri Muuga, Lisanne Knijff, and Chao Zhang from the Department of Chemistry-Ångström Laboratory, found that incorporating quadrupole moments alongside traditional dipole moments considerably enhances the performance of machine learning models. This breakthrough has important implications for fields such as drug discovery and materials science, where rapid and precise calculations of molecular interactions are crucial.
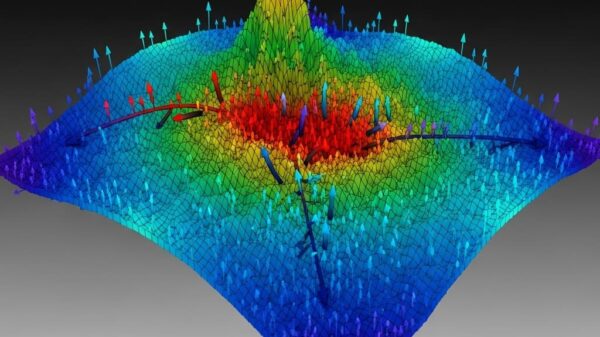
The researchers developed a convolutional neural network architecture known as PiNet2, which was specifically designed to utilize dipole and quadrupole moments to infer MEP. Their findings, validated through extensive testing on the QM9 and SPICE datasets, show that models leveraging quadrupole contributions significantly outperform those relying solely on dipole moments. This improvement highlights the pivotal role that quadrupole moments can play in the development of computational methods aimed at exploring a wider range of chemical environments.
In addition to the PiNet2 model, the research introduces PhysNet, a novel machine learning framework capable of predicting multiple molecular properties directly from atomic structures. Unlike traditional approaches that require separate post-processing for various properties, PhysNet can simultaneously predict energies, atomic forces, dipole moments, and partial charges. This integrated methodology marks a significant advancement in the application of machine learning to molecular modeling and offers a more comprehensive understanding of molecular interactions.
Both PiNet2 and PhysNet were trained and validated using the QM9 dataset, which contains approximately 134,000 small organic molecules with quantum-mechanically computed properties, as well as the larger SPICE dataset, which includes over 270,000 entries of drug-like molecules and peptides relevant for biomolecular applications. The models were optimized using high-quality reference data generated from density functional theory calculations, specifically utilizing the ωB97M-V functional with the def2-SVP basis set.
PhysNet demonstrated state-of-the-art accuracy in predicting various molecular properties, achieving highly competitive results on both datasets. Its ability to predict dipole moments and atomic charges, which are often challenging for machine learning models, stands out as a particular strength. Furthermore, PhysNet’s transferability ensures it generalizes effectively to molecular systems not included in the training set, a critical feature for practical applications.
The implications of this research extend beyond mere academic interest. By enhancing the prediction capabilities of molecular electrostatic potential, the methodologies developed by the Uppsala team offer a practical pathway for rapid access to MEP, essential for designing solvents and electrolytes. Future research could explore ways to refine the accuracy of both dipole and quadrupole moment predictions within the same framework, thus further optimizing the models for practical use.
In their analysis, the researchers assessed multiple PiNet2-based models, focusing on variables such as atomic charge and atomic dipole predictions. Notably, the AC-DQ model, which combines atomic charge predictions for both dipole and quadrupole moments, showed remarkable improvement in MEP recovery. The results from the SPICE 2.0 dataset corroborate the effectiveness of this model, highlighting its adaptability across diverse chemical spaces.
As the field increasingly turns to machine learning for advancements in computational chemistry, the Uppsala University researchers’ work stands as a testament to the potential for artificial intelligence to transform how scientists approach molecular modeling. The combination of innovative architectures like PiNet2 and PhysNet with the strategic inclusion of quadrupole moments addresses a critical gap in existing methodologies and paves the way for future developments in the field.
👉 More information
🗞 Molecular electrostatic potentials from machine learning models for dipole and quadrupole predictions
🧠 ArXiv: https://arxiv.org/abs/2601.10320
 AI Study Reveals Generated Faces Indistinguishable from Real Photos, Erodes Trust in Visual Media
AI Study Reveals Generated Faces Indistinguishable from Real Photos, Erodes Trust in Visual Media Gen AI Revolutionizes Market Research, Transforming $140B Industry Dynamics
Gen AI Revolutionizes Market Research, Transforming $140B Industry Dynamics Researchers Unlock Light-Based AI Operations for Significant Energy Efficiency Gains
Researchers Unlock Light-Based AI Operations for Significant Energy Efficiency Gains Tempus AI Reports $334M Earnings Surge, Unveils Lymphoma Research Partnership
Tempus AI Reports $334M Earnings Surge, Unveils Lymphoma Research Partnership Iaroslav Argunov Reveals Big Data Methodology Boosting Construction Profits by Billions
Iaroslav Argunov Reveals Big Data Methodology Boosting Construction Profits by Billions