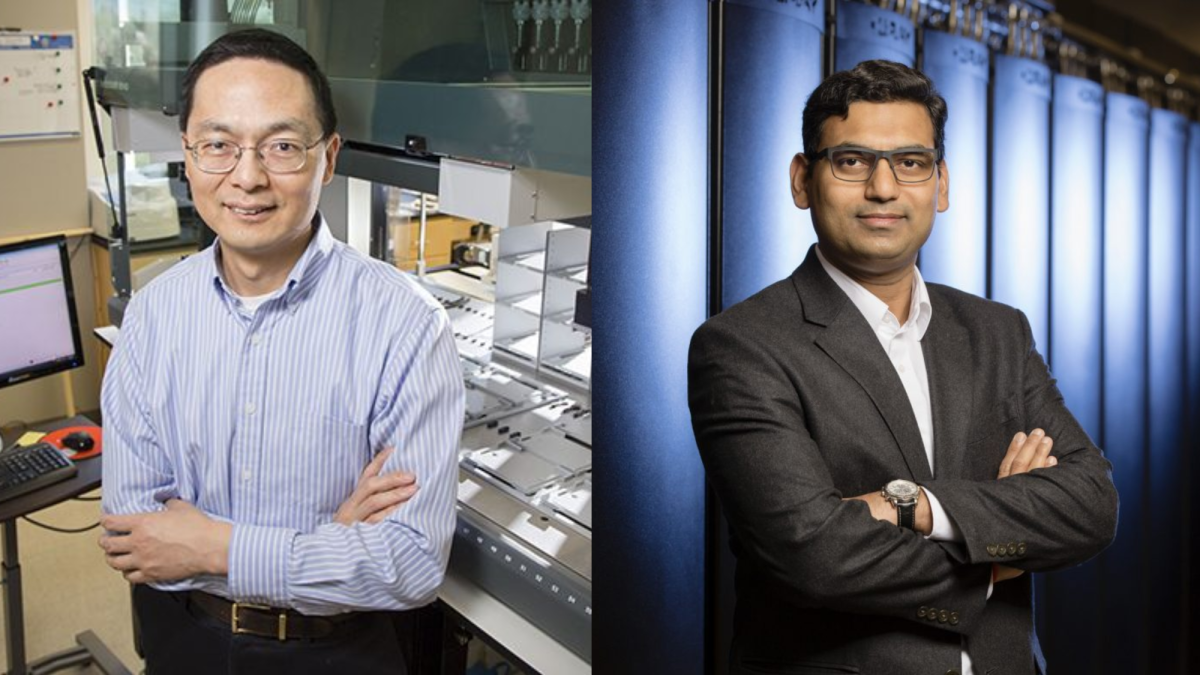
Researchers at the University have unveiled an innovative artificial intelligence model, named EZSpecificity, designed to evaluate the binding efficiency between specific enzymes and their substrates. This development, spearheaded by professors Huimin Zhao and Diwakar Shukla, is poised to accelerate advancements in drug development and synthetic biology.
Zhao, whose research focuses on leveraging synthetic biology and machine learning to explore novel chemical reaction mechanisms, collaborated with Shukla, who utilizes physics-based models to elucidate complex biological processes. Together, they aim to harness the power of machine learning—an AI subset that identifies patterns within data—to predict and optimize enzyme-substrate interactions.
Understanding EZSpecificity
Enzymes are crucial proteins that expedite chemical reactions by acting on substrates, facilitating many vital biological functions within the human body. The EZSpecificity model has the potential to illuminate pathways for developing new drugs or identifying the optimal substrates for specific enzymes. As Zhao pointed out, understanding which substrates interact effectively with particular enzymes remains a significant challenge in the field. “Inside the cell, sometimes you don’t know which enzymes really catalyze which reactions — and then we can also help to figure out the metabolism,” he explained.
The implications of better understanding enzyme-substrate relationships extend beyond drug discovery; they resonate across various domains, including enzyme engineering, synthetic biology, and biocatalysis. Shukla emphasized the labor-intensive nature of current experimentation methods, which can hinder progress: “Making the enzymes and chemicals and testing them is both tedious and precise. A highly efficient AI tool can be incredibly valuable to both researchers and the public.”
Training the Model
The EZSpecificity model was trained on two significant datasets: PDBind+ and ESIBank, which provide extensive information on enzymes, substrates, and their interactions. Shukla noted, “We tried to predict which substrate binds to which enzyme using molecular docking and simulations.” This approach enabled the creation of a comprehensive database of enzyme-substrate pairs through computational modeling, enhancing the understanding of molecular interactions.
By integrating both computational and experimental data, the model achieved a notable accuracy rate of 91.7% in identifying a single potential reactive substrate, significantly outperforming a competing model, ESP, which registered an accuracy of 58.3%.
Future Directions and Accessibility
Looking ahead, Zhao and Shukla are committed to making the EZSpecificity model accessible to the broader research community. They are developing an open-source website that will allow users to interact with the tool freely, despite holding a patent on it. Shukla stated, “Our hope is that this website will have a lot of functionalities in the future. You can come to our website and just use it open source, no restrictions.”
While the model shows promise, Zhao expressed a desire to improve its accuracy across a broader range of enzymes. “The accuracy is pretty good, more than 95%, but for other enzymes, the accuracy is low, and we do need to get more data to train the model,” he shared. Furthermore, Shukla emphasized the need to incorporate reaction kinetics into the model, stating, “If you can integrate that type of experimental and computational information into the tool, a highly accurate model for that would be very useful.”
As the field of synthetic biology continues to evolve, tools like EZSpecificity could prove invaluable in revolutionizing drug discovery and advancing our understanding of biological systems.
See also Google, Nvidia Lead $300B AI Investment Surge with Major Cloud, Chip Deals
Google, Nvidia Lead $300B AI Investment Surge with Major Cloud, Chip Deals OpenAI Enables ChatGPT Users to Disable Em Dashes with New Personalization Feature
OpenAI Enables ChatGPT Users to Disable Em Dashes with New Personalization Feature China’s Qwen AI Models Surpass US Alternatives with 385M Downloads on Hugging Face
China’s Qwen AI Models Surpass US Alternatives with 385M Downloads on Hugging Face Google Launches Autonomous Shopping Tools, Expanding Agentic Checkout and Duplex Features
Google Launches Autonomous Shopping Tools, Expanding Agentic Checkout and Duplex Features