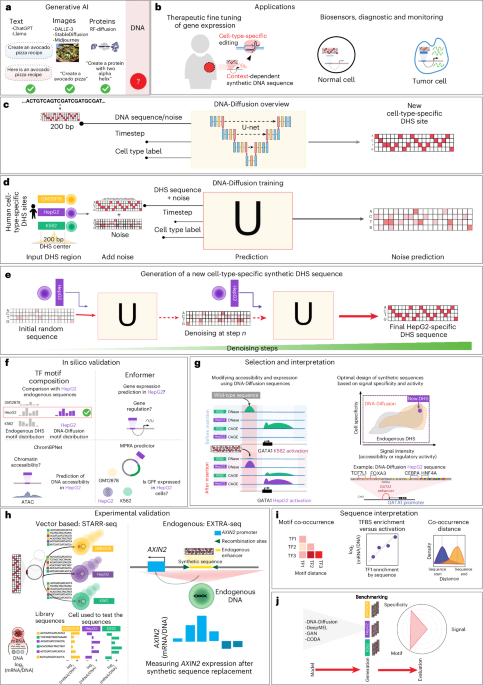
The ENCODE Project Consortium has released a comprehensive encyclopedia detailing DNA elements within the human genome, marking a significant milestone in genomic research. Published in the journal Nature, the study emphasizes the complexity of regulatory elements, presenting a cohesive framework that researchers can utilize to further understand gene expression and regulation. This work, documented in the 2012 edition of Nature, has set the stage for subsequent research aimed at unraveling the intricate dynamics of human genetics.
In a more recent development, the ENCODE Uniform Analysis Pipelines were described by Hitz et al. in a 2023 preprint on Research Square. This new approach aims to standardize data analysis across various studies, thus enhancing the reliability and reproducibility of genomic research. By facilitating seamless navigation through vast datasets, the pipelines are expected to aid researchers in deciphering the functionalities of genomic elements.
Further insights were provided by Inoue et al. in a 2017 publication, which highlighted notable differences in how enhancer activity is encoded in chromosomal versus episomal contexts. This study underscores the significance of structural variations in genomic elements and their implications in gene regulation. Such findings are essential as they guide the design of experiments aimed at manipulating enhancer functions to study their roles in various biological processes.
Looking ahead, Kagda et al. are set to release their findings in 2025, focusing on user-friendly data navigation tools within the ENCODE portal. This initiative aims to simplify access to genomic data, fostering collaboration within the scientific community. In parallel, Kundaje et al. conducted an integrative analysis of 111 reference human epigenomes, published in Nature in 2015, which provides a foundational understanding of epigenetic mechanisms driving gene regulation.
Research into specific applications of genomic data continues to expand. Martens and Stunnenberg discussed the BLUEPRINT project, which maps blood cell epigenomes, further elucidating the complexity of gene regulation in different cell types. Similar efforts have been made by Noguchi et al., who explored FANTOM5 CAGE profiles, uncovering vital data about gene expression dynamics in human and mouse samples. This work emphasizes the importance of comparative studies in understanding gene regulatory mechanisms across species.
Recent advancements in machine learning have added a new dimension to genomic research. For instance, Ho et al. described denoising diffusion probabilistic models, a technique that allows for enhanced modeling of genomic sequences. This approach, along with score-based generative modeling by Song et al., highlights the evolving interplay between artificial intelligence and genomics.
Additionally, Rombach et al. have pioneered high-resolution image synthesis with latent diffusion models, showcasing the potential for these technologies in biological imaging. The application of such models could lead to breakthroughs in visualizing complex genetic structures and their interactions.
As research progresses, the intersection of AI and genomics continues to yield innovative solutions for understanding and designing DNA sequences. The work of Li et al., who presented Diffusion-LM for controllable text generation in 2022, illuminates how these models can also influence genetic engineering, providing tools for precise manipulation of genetic sequences.
Another exciting avenue explored by Reddy et al. involves strategies for designing cell-type-specific promoter sequences using optimization techniques. This advancement may streamline the process of creating tailored genetic constructs for therapeutic applications. The synergy between computational tools and genomic research promises to revolutionize our understanding of genetic design and regulation.
In a forward-looking perspective, the cumulative efforts of these studies and initiatives signal a transformative era in genomics. The integration of advanced computational models and standardized analysis pipelines not only enhances our understanding of the human genome but also paves the way for innovative therapeutic strategies. As researchers continue to unravel the complexities of DNA, the potential for groundbreaking discoveries in medicine and biotechnology becomes ever more tangible.
See also DuckDuckGo Launches AI Image Generator with Enhanced Privacy Features
DuckDuckGo Launches AI Image Generator with Enhanced Privacy Features Intel Launches $300 B50 GPU Offering Solid Performance for AI and 3D Tasks in 2025
Intel Launches $300 B50 GPU Offering Solid Performance for AI and 3D Tasks in 2025 AI Study Reveals Image Generation Converges to 12 Styles After 100 Cycles
AI Study Reveals Image Generation Converges to 12 Styles After 100 Cycles China Files 700 Generative AI Models as HarmonyOS Devices Exceed 1.19 Billion Units
China Files 700 Generative AI Models as HarmonyOS Devices Exceed 1.19 Billion Units