February 21, 2026
3 min read
Add Us On GoogleAdd SciAm
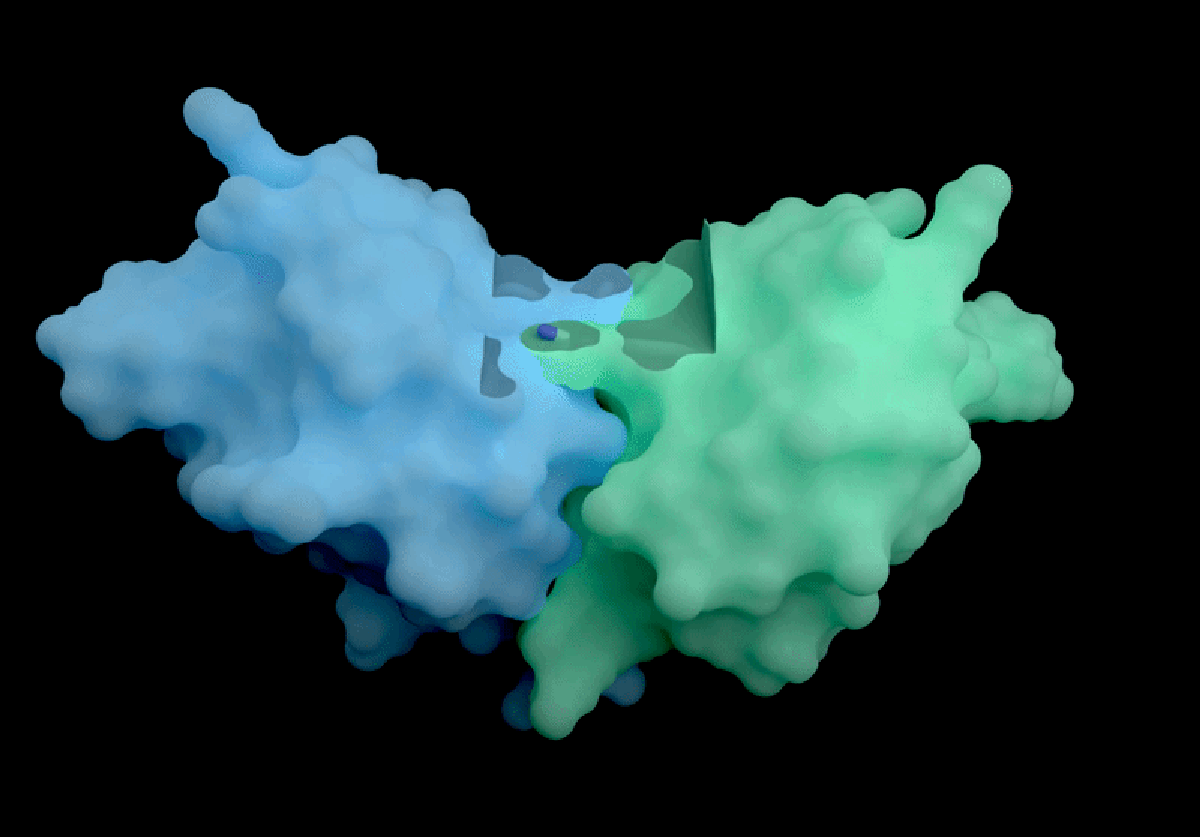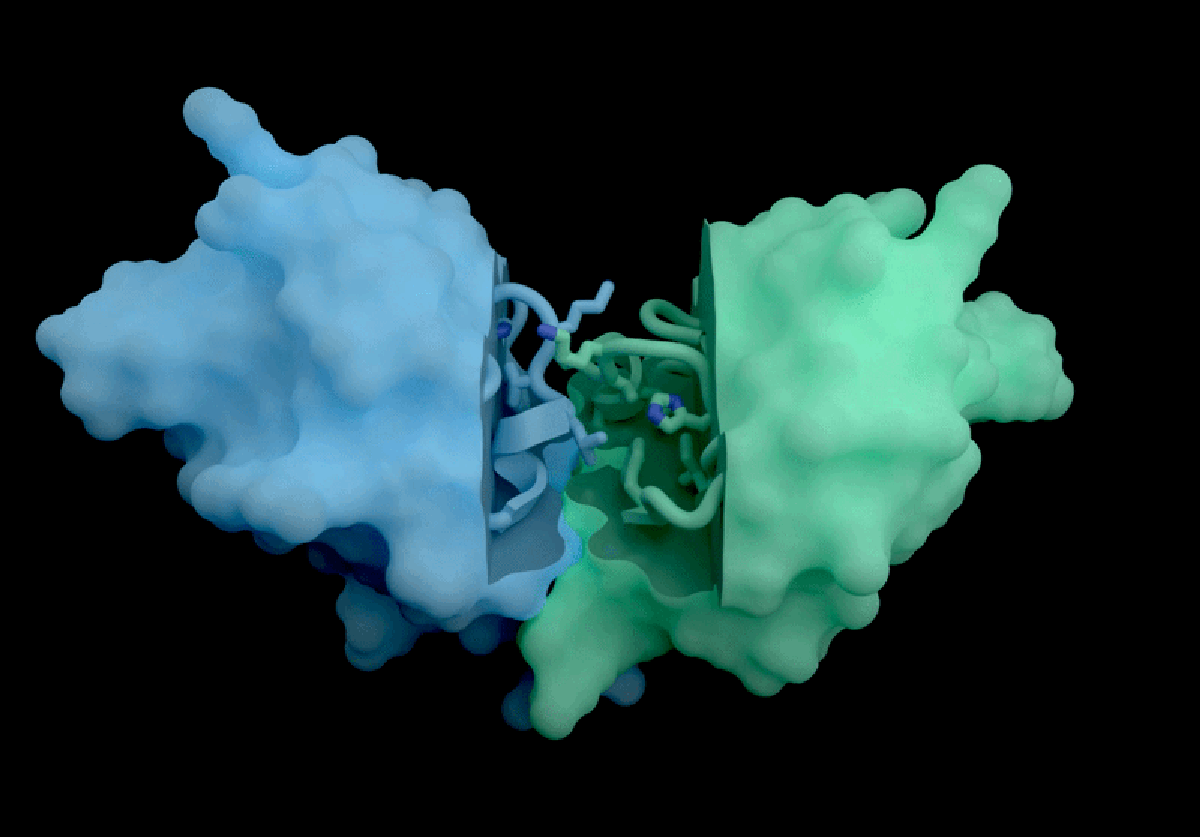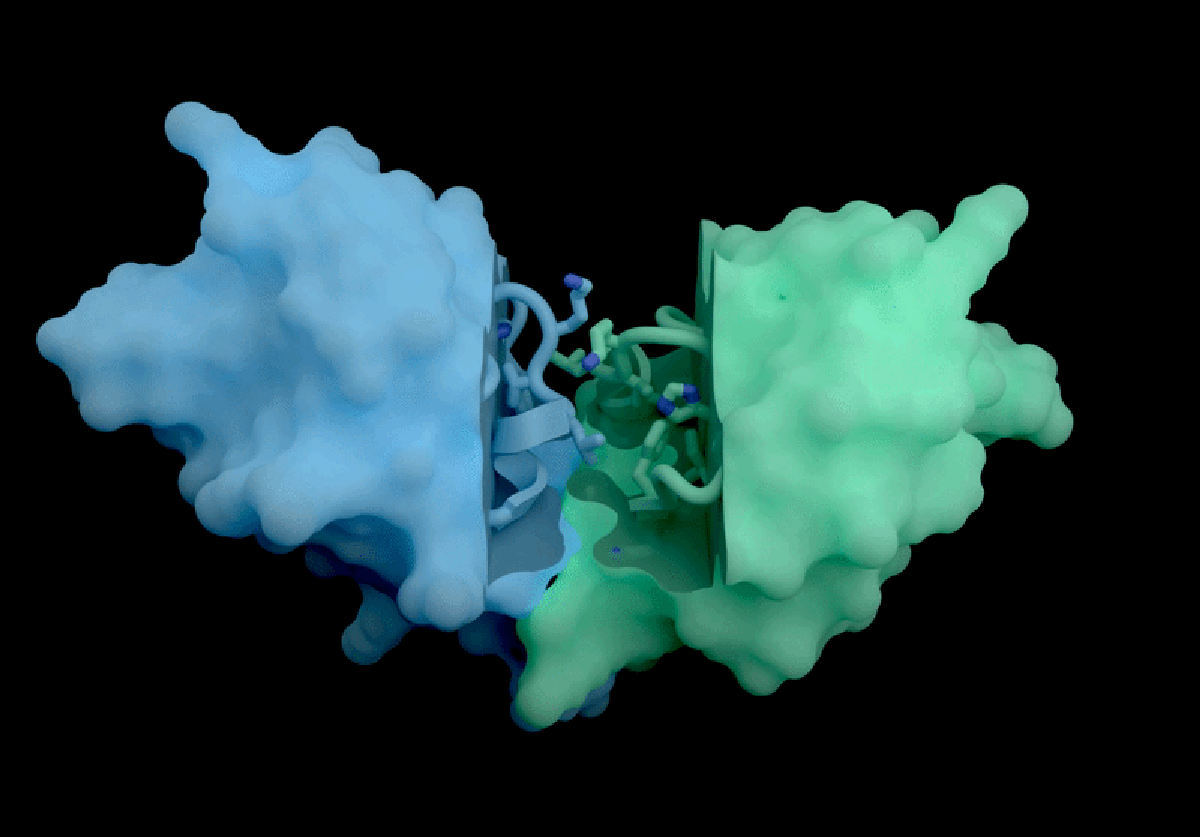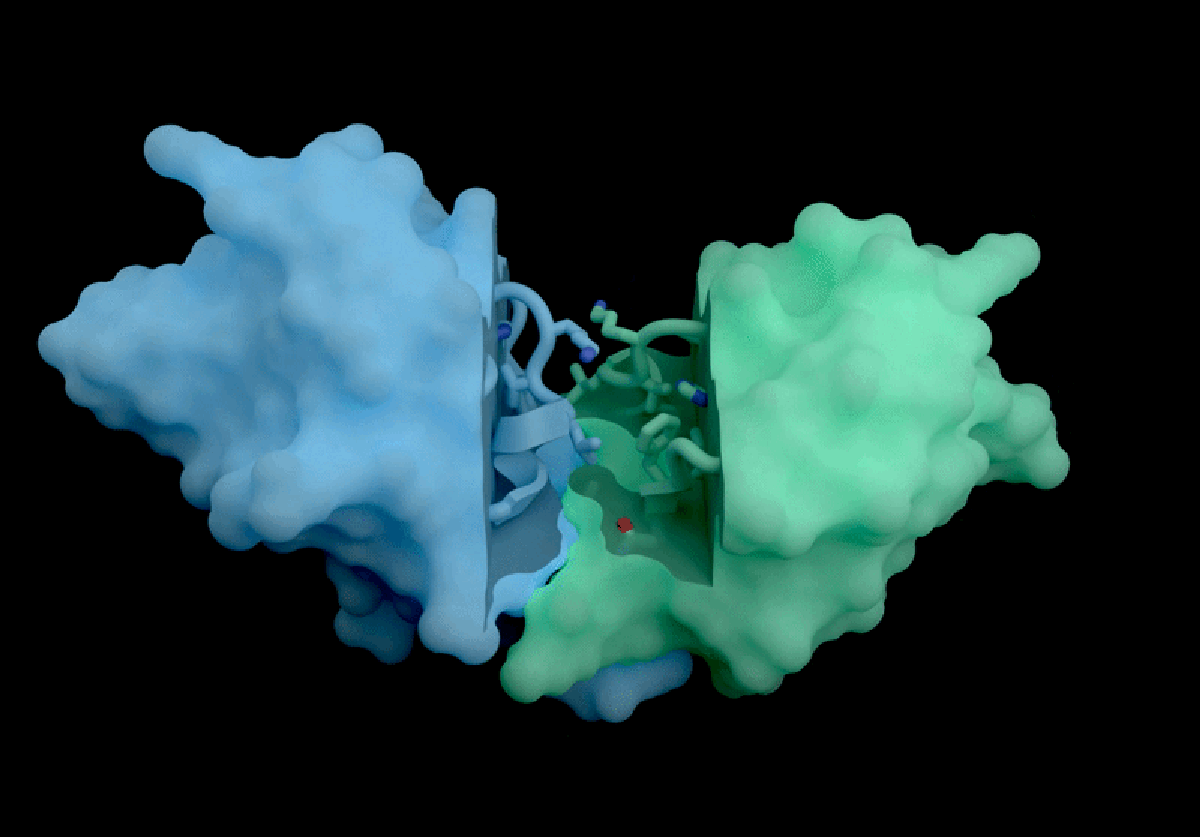
Isomorphic Labs, a spin-off from Google DeepMind, has introduced a groundbreaking artificial intelligence model called IsoDDE, which promises significant advancements in drug discovery, as detailed in a technical report released on February 10, 2026. The London-based company claims that the IsoDDE model can accurately predict how proteins interact with potential therapeutic molecules, marking a notable evolution from its predecessor, AlphaFold3, which was launched nearly two years ago.
While AlphaFold3 was designed with drug discovery in mind, the new IsoDDE model has reportedly surpassed its capabilities. This proprietary tool, however, remains under wraps, raising concerns among researchers who have been developing open-source alternatives. Unlike the widely available AlphaFold AI systems, which have significantly contributed to the scientific community, IsoDDE’s details are closely guarded, limiting insights that other researchers could utilize to replicate or build upon its performance.
“It’s a major advance, on the scale of an AlphaFold4,” noted Mohammed AlQuraishi, a computational biologist at Columbia University, who is actively engaged in creating fully open-source versions of AlphaFold. AlQuraishi expressed frustration over the lack of transparency, stating that “the problem is that we know nothing of the details.”
IsoDDE’s performance includes impressive capabilities, such as predicting binding affinity and interactions between potential drugs and proteins, a crucial factor in the development of new therapeutics. The report states that IsoDDE outperforms both Boltz-2, an open-source model developed by the Massachusetts Institute of Technology, and traditional physics-based methods used for determining binding affinities.
AlQuraishi praised IsoDDE for its ability to predict drug–protein interactions, even with molecules that significantly differ from the training data. “That’s the really hard problem, and suggests that they must’ve done something pretty novel,” he remarked.
Max Jaderberg, president of Isomorphic Labs, emphasized that the models behind IsoDDE are “profoundly different” from existing efforts. However, he did not disclose any specifics regarding the underlying algorithms or data strategies employed. Jaderberg stated that “it’s a combination of compute, data, and algorithms,” and expressed hope that the report would inspire other teams to advance their drug-discovery AIs.
The development of IsoDDE has coincided with lucrative partnerships with major pharmaceutical companies such as Johnson & Johnson, Eli Lilly, and Novartis, potentially worth billions of pounds. Isomorphic also has an internal pipeline for drug development, with clinical trials anticipated in the near future.
As the field of AI-driven drug discovery evolves, it raises questions about the balance between proprietary technology and the need for open-source collaboration in scientific research. The proprietary nature of IsoDDE may pose challenges for researchers aiming to replicate its success using publicly available data. However, experts like Gabriele Corso, a machine-learning scientist who co-developed Boltz-2, argue that many improvements can still be made with existing data. “I think this is a new baseline to match—but also to pass,” he asserted.
This development signals a pivotal moment in the intersection of artificial intelligence and pharmaceuticals, highlighting the potential of proprietary AI models while also emphasizing the importance of transparency and collaboration in advancing scientific research.
See also AI Transforms Health Care Workflows, Elevating Patient Care and Outcomes
AI Transforms Health Care Workflows, Elevating Patient Care and Outcomes Tamil Nadu’s Anbil Mahesh Seeks Exemption for In-Service Teachers from TET Requirements
Tamil Nadu’s Anbil Mahesh Seeks Exemption for In-Service Teachers from TET Requirements Top AI Note-Taking Apps of 2026: Boost Productivity with 95% Accurate Transcriptions
Top AI Note-Taking Apps of 2026: Boost Productivity with 95% Accurate Transcriptions